Note
Go to the end to download the full example code.
Using scikit-matter selectors with scikit-learn pipelines#
import numpy as np
from matplotlib import pyplot as plt
from sklearn.datasets import load_diabetes
from sklearn.linear_model import RidgeCV
from sklearn.model_selection import train_test_split
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler
from skmatter.feature_selection import CUR, FPS
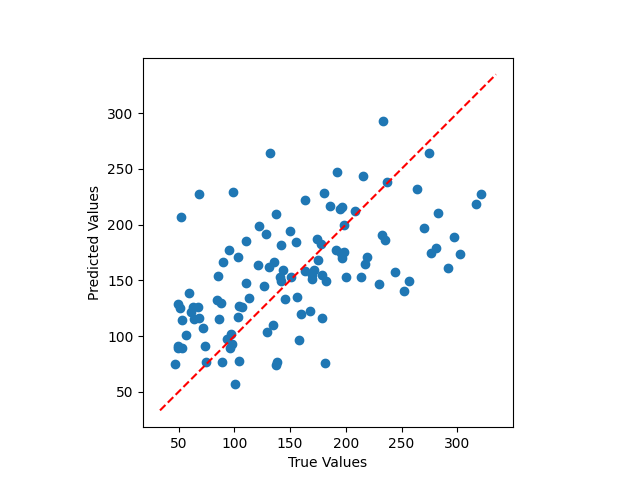
Simple integration of scikit-matter selectors#
This example shows how to use FPS to subselect features before training a RidgeCV.
scaler = StandardScaler()
selector = FPS(n_to_select=4)
ridge = RidgeCV(cv=2, alphas=np.logspace(-8, 2, 10))
X, y = load_diabetes(return_X_y=True)
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=0)
pipe = Pipeline([("scaler", scaler), ("selector", selector), ("ridge", ridge)])
pipe.fit(X_train.copy(), y_train.copy())
plt.scatter(y_test, pipe.predict(X_test))
plt.gca().set_aspect("equal")
plt.plot(plt.xlim(), plt.xlim(), "r--")
plt.xlabel("True Values")
plt.ylabel("Predicted Values")
plt.show()
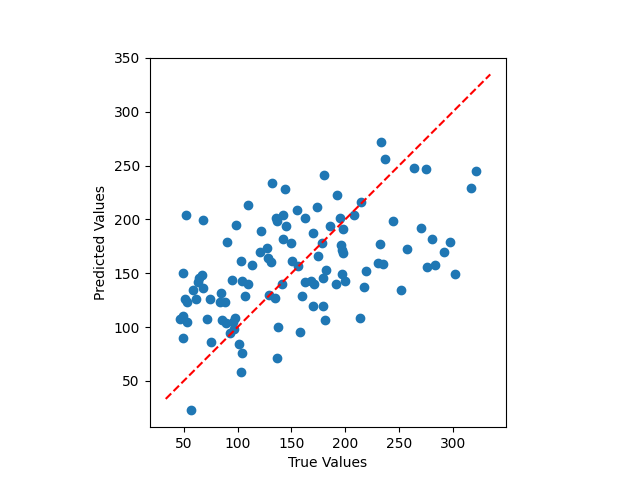
Stacking selectors one after another#
This example shows how to use an FPS, then CUR selector to subselect features before training a RidgeCV.
scaler = StandardScaler()
fps = FPS(n_to_select=8)
cur = CUR(n_to_select=4)
ridge = RidgeCV(cv=2, alphas=np.logspace(-8, 2, 10))
X, y = load_diabetes(return_X_y=True)
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=0)
pipe = Pipeline(
[("scaler", scaler), ("selector1", fps), ("selector2", cur), ("ridge", ridge)]
)
pipe.fit(X_train.copy(), y_train.copy())
plt.scatter(y_test, pipe.predict(X_test))
plt.gca().set_aspect("equal")
plt.plot(plt.xlim(), plt.xlim(), "r--")
plt.xlabel("True Values")
plt.ylabel("Predicted Values")
plt.show()