Note
Go to the end to download the full example code.
KPCovC Hyperparameter Tuning#
import matplotlib as mpl
import matplotlib.pyplot as plt
from sklearn import datasets
from sklearn.preprocessing import StandardScaler
from sklearn.decomposition import KernelPCA
from skmatter.decomposition import KernelPCovC
plt.rcParams["image.cmap"] = "tab20"
plt.rcParams["scatter.edgecolors"] = "k"
random_state = 0
n_components = 2
For this, we will use the sklearn.datasets.make_circles() dataset from
sklearn.
X, y = datasets.make_circles(
noise=0.1, factor=0.7, random_state=random_state, n_samples=1500
)
Original Data#
fig, ax = plt.subplots(figsize=(5.5, 5))
ax.scatter(X[:, 0], X[:, 1], c=y)
ax.set_title("Original Data")
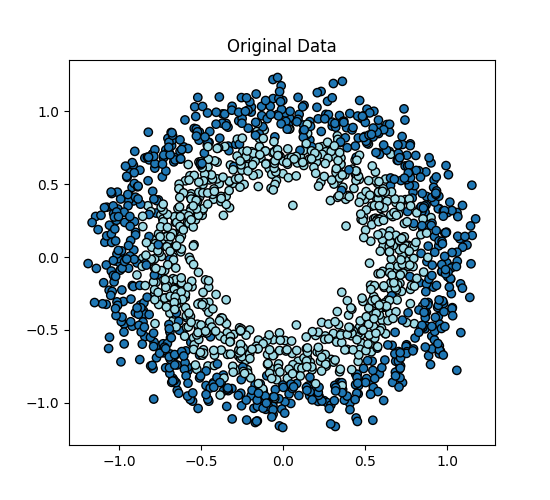
Text(0.5, 1.0, 'Original Data')
Scale Data
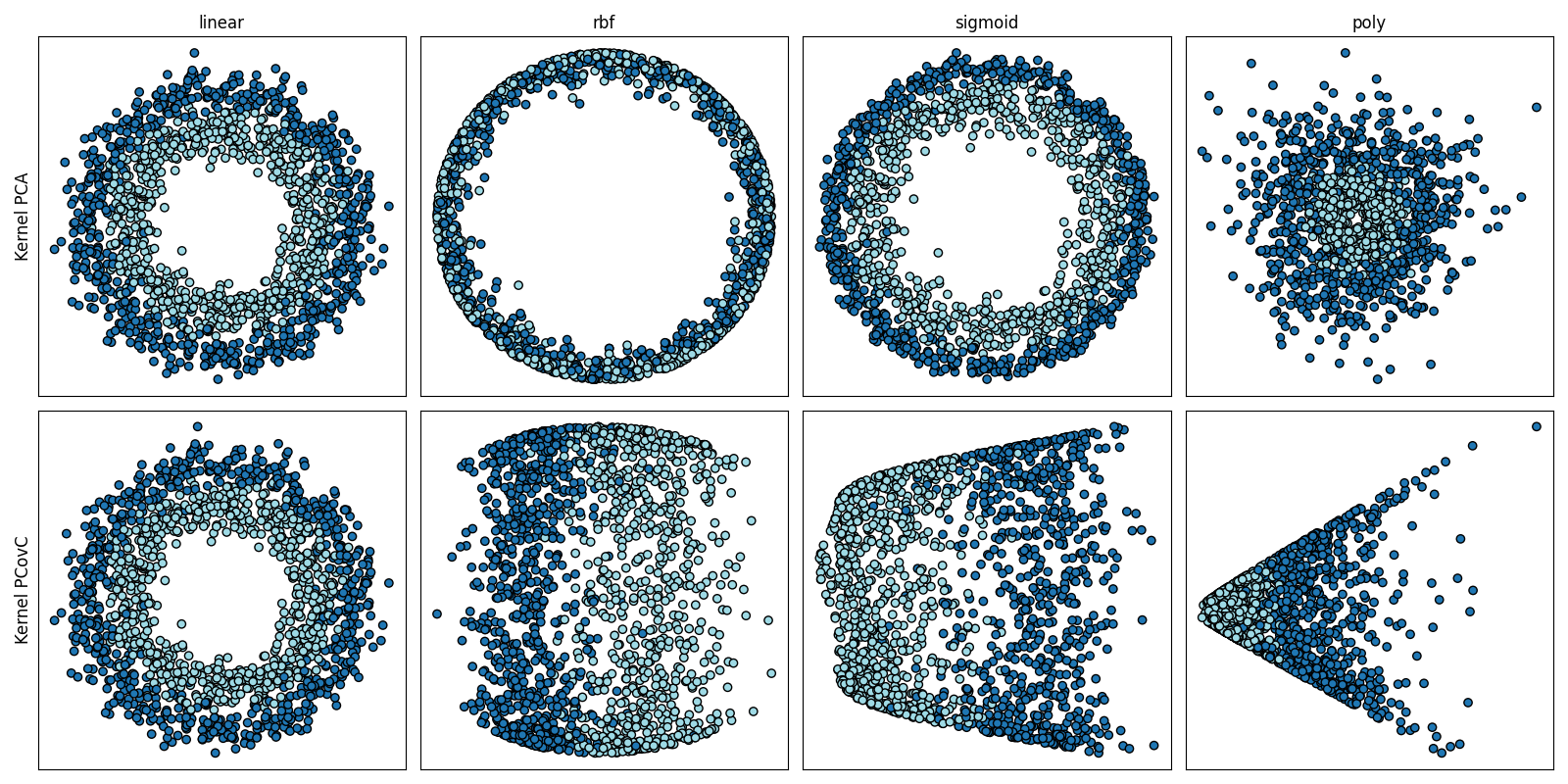
Effect of Kernel on KPCA and KPCovC Projections#
Here, we see how Kernel PCovC with kernels such as a radial basis function can outperform Kernel PCA by producing cleanly separable projections from noisy circular data.
kernels = ["linear", "rbf", "sigmoid", "poly"]
kernel_params = {
"rbf": {"gamma": 0.5},
"sigmoid": {"gamma": 1.0},
"poly": {"degree": 6},
}
fig, axs = plt.subplots(2, len(kernels), figsize=(len(kernels) * 4, 8))
center = True
mixing = 0.10
for i, kernel in enumerate(kernels):
kpca = KernelPCA(
random_state=random_state,
n_components=n_components,
kernel=kernel,
**kernel_params.get(kernel, {}),
)
t_kpca = kpca.fit_transform(X_scaled)
kpcovc = KernelPCovC(
n_components=n_components,
mixing=mixing,
kernel=kernel,
random_state=random_state,
**kernel_params.get(kernel, {}),
center=center,
)
t_kpcovc = kpcovc.fit_transform(X_scaled, y)
axs[0][i].scatter(t_kpca[:, 0], t_kpca[:, 1], c=y)
axs[1][i].scatter(t_kpcovc[:, 0], t_kpcovc[:, 1], c=y)
axs[0][i].set_title(kernel)
axs[0][i].set_xticks([])
axs[1][i].set_xticks([])
axs[0][i].set_yticks([])
axs[1][i].set_yticks([])
axs[0][0].set_ylabel("Kernel PCA", fontsize=mpl.rcParams["axes.titlesize"])
axs[1][0].set_ylabel("Kernel PCovC", fontsize=mpl.rcParams["axes.titlesize"])
fig.subplots_adjust(wspace=0, hspace=0)
plt.tight_layout()
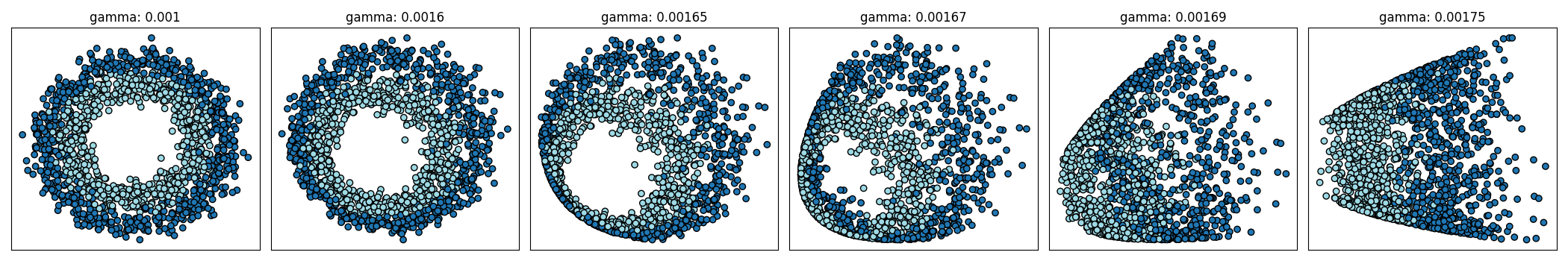
Decision Boundary Formation with Gamma Tuning#
Depending on the data, tuning gamma values for the KPCovC kernel can greatly improve latent space projections, enabling clearer decision boundaries.
gamma_vals = [0.001, 0.0016, 0.00165, 0.00167, 0.00169, 0.00175]
fig, axs = plt.subplots(1, len(gamma_vals), figsize=(len(gamma_vals) * 3.5, 3.5))
for ax, gamma in zip(axs, gamma_vals):
kpcovc = KernelPCovC(
n_components=n_components,
random_state=random_state,
mixing=mixing,
center=center,
kernel="rbf",
gamma=gamma,
)
t_kpcovc = kpcovc.fit_transform(X_scaled, y)
ax.scatter(t_kpcovc[:, 0], t_kpcovc[:, 1], c=y)
ax.set_title(f"gamma: {gamma}")
ax.set_xticks([])
ax.set_yticks([])
fig.subplots_adjust(wspace=0)
plt.tight_layout()