Note
Go to the end to download the full example code.
Feature Selection on the WHO Dataset#
import numpy as np
import scipy
from matplotlib import pyplot as plt
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import train_test_split
from skmatter.datasets import load_who_dataset
from skmatter.feature_selection import CUR, FPS, PCovCUR, PCovFPS
from skmatter.preprocessing import StandardFlexibleScaler
Load the Dataset#
df = load_who_dataset()["data"]
print(df)
Country Year ... SN.ITK.DEFC.ZS NY.GDP.PCAP.CD
0 Afghanistan 2005 ... 36.1 255.055120
1 Afghanistan 2006 ... 33.3 274.000486
2 Afghanistan 2007 ... 29.8 375.078128
3 Afghanistan 2008 ... 26.5 387.849174
4 Afghanistan 2009 ... 23.3 443.845151
... ... ... ... ... ...
2015 South Africa 2015 ... 5.2 6204.929901
2016 South Africa 2016 ... 5.4 5735.066787
2017 South Africa 2017 ... 5.5 6734.475153
2018 South Africa 2018 ... 5.7 7048.522211
2019 South Africa 2019 ... 6.3 6688.787271
[2020 rows x 12 columns]
columns = np.array(
[
"SP.POP.TOTL",
"SH.TBS.INCD",
"SH.IMM.MEAS",
"SE.XPD.TOTL.GD.ZS",
"SH.DYN.AIDS.ZS",
"SH.IMM.IDPT",
"SH.XPD.CHEX.GD.ZS",
"SN.ITK.DEFC.ZS",
"NY.GDP.PCAP.CD",
]
)
column_names = np.array(
[
"Population",
"Tuberculosis",
"Immunization, measles",
"Educ. Expenditure",
"HIV",
"Immunization, DPT",
"Health Expenditure",
"Undernourishment",
"GDP per capita",
]
)
columns = columns[[8, 4, 2, 6, 1, 7, 0, 5, 3]].tolist()
column_names = column_names[[8, 4, 2, 6, 1, 7, 0, 5, 3]].tolist()
We are taking the logarithm of the population and GDP to avoid extreme distributions
log_scaled = ["SP.POP.TOTL", "NY.GDP.PCAP.CD"]
for ls in log_scaled:
print(X_raw[:, columns.index(ls)].min(), X_raw[:, columns.index(ls)].max())
if ls in columns:
X_raw[:, columns.index(ls)] = np.log10(X_raw[:, columns.index(ls)])
y_raw = np.array(df["SP.DYN.LE00.IN"]) # [np.where(df['Year']==2000)[0]])
y_raw = y_raw.reshape(-1, 1)
X_raw.shape
149841.0 7742681934.0
110.460874721483 123678.70214327476
(2020, 9)
Scale and Center the Features and Targets#
x_scaler = StandardFlexibleScaler(column_wise=True)
X = x_scaler.fit_transform(X_raw)
y_scaler = StandardFlexibleScaler(column_wise=True)
y = y_scaler.fit_transform(y_raw)
n_components = 2
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, shuffle=True, random_state=0
)
Provide an estimated target for the feature selector#
kernel_params = {"kernel": "rbf", "gamma": 0.08858667904100832}
lr = LinearRegression(fit_intercept=False)
yp_train = lr.fit(X_train, y_train).predict(X_train)
Compute the Selections for Each Selector Type#
PCov-CUR#
0%| | 0/9 [00:00<?, ?it/s]
100%|██████████| 9/9 [00:00<00:00, 229.67it/s]
PCov-FPS#
0%| | 0/8 [00:00<?, ?it/s]
100%|██████████| 8/8 [00:00<00:00, 9391.11it/s]
CUR#
0%| | 0/9 [00:00<?, ?it/s]
100%|██████████| 9/9 [00:00<00:00, 952.46it/s]
FPS#
0%| | 0/8 [00:00<?, ?it/s]
100%|██████████| 8/8 [00:00<00:00, 8719.97it/s]
(For Comparison) Recursive Feature Addition#
class RecursiveFeatureAddition:
"""Class for recursive feature addition"""
def __init__(self, n_to_select):
"""Init."""
self.n_to_select = n_to_select
self.selected_idx_ = np.zeros(n_to_select, dtype=int)
def fit(self, X, y):
"""Perform the fit."""
remaining = np.arange(X.shape[1])
for n in range(self.n_to_select):
errors = np.zeros(len(remaining))
for i, pp in enumerate(remaining):
lr.fit(X[:, [*self.selected_idx_[:n], pp]], y)
errors[i] = lr.score(X[:, [*self.selected_idx_[:n], pp]], y)
self.selected_idx_[n] = remaining[np.argmax(errors)]
remaining = np.array(np.delete(remaining, np.argmax(errors)), dtype=int)
return self
rfa = RecursiveFeatureAddition(n_select).fit(X_train, y_train)
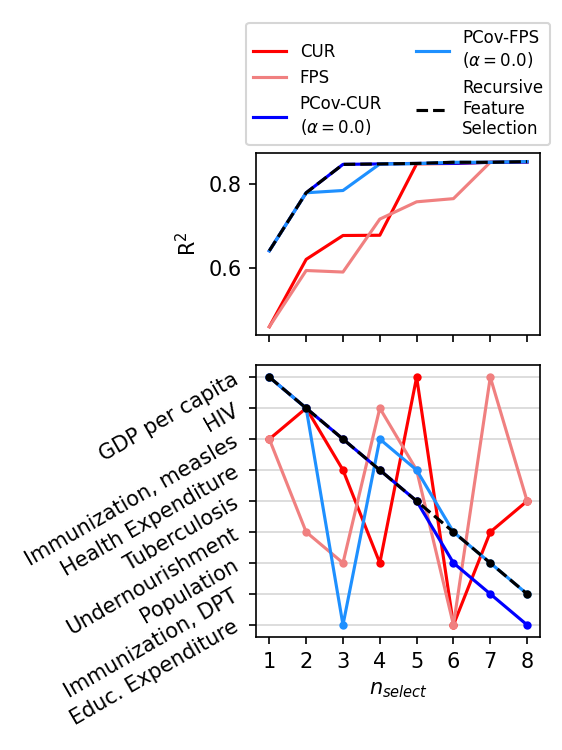
Plot our Results#
fig, axes = plt.subplots(
2,
1,
figsize=(3.75, 5),
gridspec_kw=dict(height_ratios=(1, 1.5)),
sharex=True,
dpi=150,
)
ns = np.arange(1, n_select, dtype=int)
all_errors = {}
for selector, color, linestyle, label in zip(
[cur, fps, pcur, pfps, rfa],
["red", "lightcoral", "blue", "dodgerblue", "black"],
["solid", "solid", "solid", "solid", "dashed"],
[
"CUR",
"FPS",
"PCov-CUR\n" + r"($\alpha=0.0$)",
"PCov-FPS\n" + r"($\alpha=0.0$)",
"Recursive\nFeature\nSelection",
],
):
if label not in all_errors:
errors = np.zeros(len(ns))
for i, n in enumerate(ns):
lr.fit(X_train[:, selector.selected_idx_[:n]], y_train)
errors[i] = lr.score(X_test[:, selector.selected_idx_[:n]], y_test)
all_errors[label] = errors
axes[0].plot(ns, all_errors[label], c=color, label=label, linestyle=linestyle)
axes[1].plot(
ns, selector.selected_idx_[: max(ns)], c=color, marker=".", linestyle=linestyle
)
axes[1].set_xlabel(r"$n_{select}$")
axes[1].set_xticks(range(1, n_select))
axes[0].set_ylabel(r"R$^2$")
axes[1].set_yticks(np.arange(X.shape[1]))
axes[1].set_yticklabels(column_names, rotation=30, fontsize=10)
axes[0].legend(ncol=2, fontsize=8, bbox_to_anchor=(0.5, 1.0), loc="lower center")
axes[1].invert_yaxis()
axes[1].grid(axis="y", alpha=0.5)
plt.tight_layout()
plt.show()
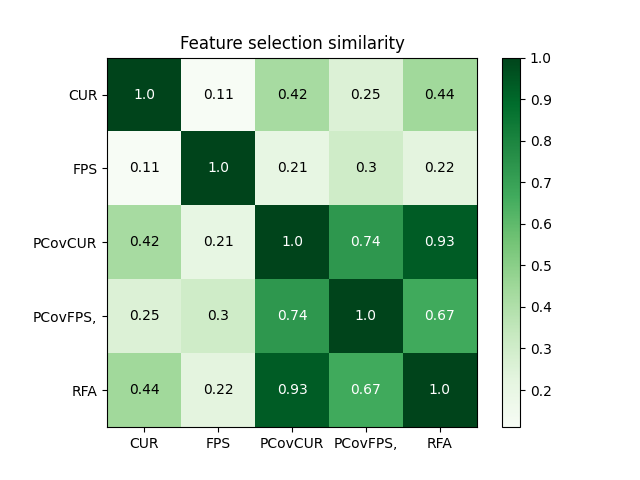
Plot correlation between selectors#
selected_idx = np.array(
[selector.selected_idx_ for selector in [cur, fps, pcur, pfps, rfa]]
).T
similarity = np.zeros((len(selected_idx.T), len(selected_idx.T)))
for i in range(len(selected_idx.T)):
for j in range(len(selected_idx.T)):
similarity[i, j] = scipy.stats.weightedtau(
selected_idx[:, i], selected_idx[:, j], rank=False
)[0]
labels = ["CUR", "FPS", "PCovCUR", "PCovFPS,", "RFA"]
plt.imshow(similarity, cmap="Greens")
plt.xticks(np.arange(len(labels)), labels=labels)
plt.yticks(np.arange(len(labels)), labels=labels)
plt.title("Feature selection similarity")
for i in range(len(labels)):
for j in range(len(labels)):
value = np.round(similarity[i, j], 2)
color = "white" if value > 0.5 else "black"
text = plt.gca().text(j, i, value, ha="center", va="center", color=color)
plt.colorbar()
plt.show()