Note
Go to the end to download the full example code.
PCovC Hyperparameter Tuning#
import matplotlib.pyplot as plt
from matplotlib.colors import LinearSegmentedColormap
from sklearn.datasets import load_iris
from sklearn.decomposition import PCA
from sklearn.inspection import DecisionBoundaryDisplay
from sklearn.linear_model import LogisticRegressionCV, Perceptron, RidgeClassifierCV
from sklearn.preprocessing import StandardScaler
from sklearn.svm import LinearSVC
from skmatter.decomposition import PCovC
plt.rcParams["image.cmap"] = "tab10"
plt.rcParams["scatter.edgecolors"] = "k"
random_state = 10
n_components = 2
For this, we will use the sklearn.datasets.load_iris() dataset from
sklearn.
X, y = load_iris(return_X_y=True)
scaler = StandardScaler()
X_scaled = scaler.fit_transform(X)
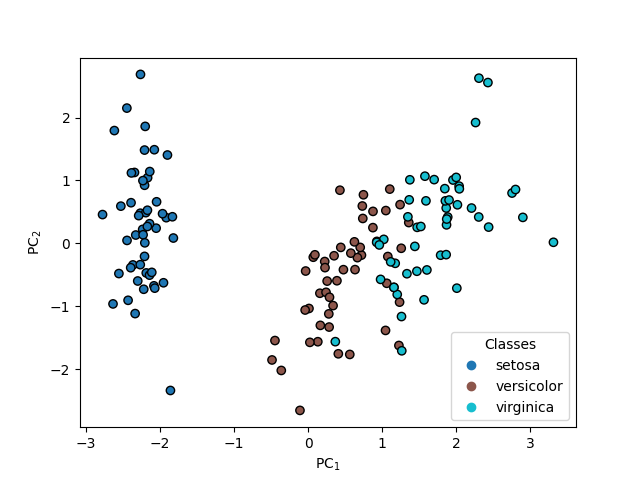
PCA#
pca = PCA(n_components=n_components)
pca.fit(X_scaled, y)
T_pca = pca.transform(X_scaled)
fig, axis = plt.subplots()
scatter = axis.scatter(T_pca[:, 0], T_pca[:, 1], c=y)
axis.set(xlabel="PC$_1$", ylabel="PC$_2$")
axis.legend(
scatter.legend_elements()[0],
load_iris().target_names,
loc="lower right",
title="Classes",
)
<matplotlib.legend.Legend object at 0x7ee561fb0e10>
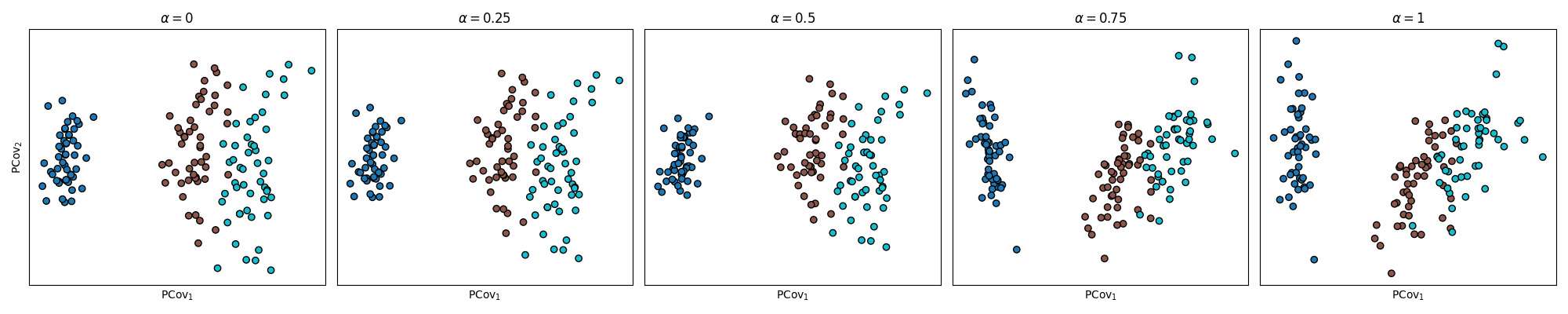
Effect of Mixing Parameter \(\alpha\) on PCovC Map#
Below, we see how different \(\alpha\) values for our PCovC model result in varying class distinctions between setosa, versicolor, and virginica on the PCovC map.
n_mixing = 5
mixing_params = [0, 0.25, 0.50, 0.75, 1]
fig, axs = plt.subplots(1, n_mixing, figsize=(4 * n_mixing, 4), sharey="row")
for ax, mixing in zip(axs, mixing_params):
pcovc = PCovC(
mixing=mixing,
n_components=n_components,
random_state=random_state,
classifier=LogisticRegressionCV(),
)
pcovc.fit(X_scaled, y)
T = pcovc.transform(X_scaled)
ax.set_xticks([])
ax.set_yticks([])
ax.set_title(r"$\alpha=$" + str(mixing))
ax.set_xlabel("PCov$_1$")
ax.scatter(T[:, 0], T[:, 1], c=y)
axs[0].set_ylabel("PCov$_2$")
fig.subplots_adjust(wspace=0)
plt.tight_layout()
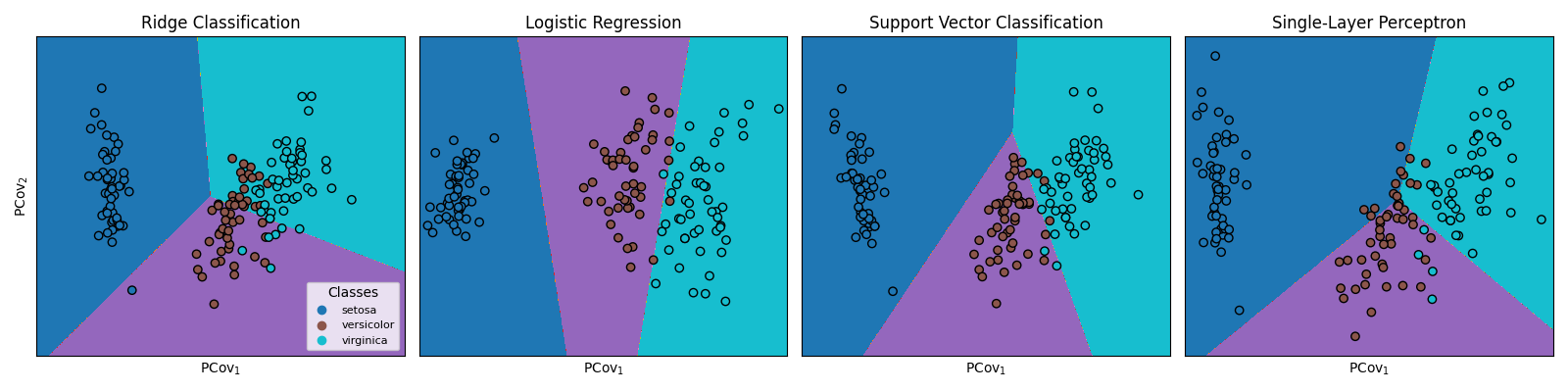
Effect of PCovC Classifier on PCovC Maps and Decision Boundaries#
Here, we see how a PCovC model (\(\alpha\) = 0.5) fitted with different classifiers produces varying PCovC maps. In addition, we see the varying decision boundaries produced by the respective PCovC classifiers.
mixing = 0.5
fig, axs = plt.subplots(1, 4, figsize=(16, 4))
models = {
RidgeClassifierCV(): "Ridge Classification",
LogisticRegressionCV(random_state=random_state): "Logistic Regression",
LinearSVC(random_state=random_state): "Support Vector Classification",
Perceptron(random_state=random_state): "Single-Layer Perceptron",
}
for ax, model in zip(axs, models):
pcovc = PCovC(
mixing=mixing,
n_components=n_components,
random_state=random_state,
classifier=model,
)
pcovc.fit(X_scaled, y)
T = pcovc.transform(X_scaled)
ax.set_title(models[model])
DecisionBoundaryDisplay.from_estimator(
estimator=pcovc.classifier_,
X=T,
ax=ax,
response_method="predict",
grid_resolution=1000,
)
scatter = ax.scatter(T[:, 0], T[:, 1], c=y)
ax.set_xlabel("PCov$_1$")
ax.set_xticks([])
ax.set_yticks([])
axs[0].set_ylabel("PCov$_2$")
axs[0].legend(
scatter.legend_elements()[0],
load_iris().target_names,
loc="lower right",
title="Classes",
fontsize=8,
)
fig.subplots_adjust(wspace=0.04)
plt.tight_layout()
plt.show()